Diseases
Coronaviruses in pigs
Chapter select
Control Tools
-
Diagnostics availability
-
Commercial diagnostic kits available worldwide
Commercial test kits that are available for CoV in pigs include antibody detection via ELISAs, RNA detection via PCR and antigen or RNA detection via lateral flow devices. Due the similarity of TGEV/PRCV, often commercial kits are combined to detect both and differentiate between them.
Nucleic acid detection (faeces or intestinal samples): RT-PCR or RT-real-time PCR kits:
- TGEV
- TGEV/PRCV differential kits
- PEDV
- PEDV/TGEV
- PEDV/TGEV/PDCoV differential kits
- PDCoV
- PEDV/PDCoV
- pHEV
Antigen detection in fixed intestinal tissues or lungs:
- TGEV/PRCV, cross-reaction, no differentiation
- PEDV
Antibody detection (serum):
- Blocking ELISA
- TGEV
- TGEV/PRCV; differentiation possible, cross reactivity a common problem
- Indirect ELISA
- PEDV
- PEDV IgA (China only, colostrum and serum)
- PEDV FA Substrate slides
Neutralization antibody detection (serum):Offered by some specialized laboratories; requires the ability to grow the virus.
- PEDV
List of commercially available tests (Diagnostics for Animals)
GAPS :
- Most commercial serology assays (ELISAs) are based on utilizing serum samples. There is a need to also investigate other sample types such as enteric content, oral fluid or milk/colostrum. Most assays are either blocking assays (no isotype differentiation) or based on IgG detection. Validated and commercial assays would be beneficial for IgA and IgM assessment.
- No “on farm” commercial lateral assays appear to be available. Only experimental protocols have been published.
- A more standardized validation approach including ring assays would be beneficial to be aware of differences in sensitivity and specificity among tests.
- No commercial assays are currently in place to test pigs for SeACoV or PHEV. Especially for PHEV, in house assays are very difficult to achieve as they often require viable virus and primary cells for culture.
- While there is a commercial PCR assay in place for PDCoV, having a serology assay may also be useful as it is known that shedding of PDCoV is short and the disease, if present on a farm, may be missed.
-
Commercial diagnostic kits available in Europe
Same as above but no IgA ELISA. List of commercially available tests (Diagnostics for Animals)
GAPS :
- More on-farm tests may be needed and also a better ability to multiplex numerous if not all CoV targets in a kit. This would reduce the overall cost for farmers and allow to detect the introduction of any unexpected CoV in real-time.
- In the past some CoV were routinely screened for in meat-juices obtained from routine abattoir collections. These were used mainly for antibody assays (TGEV). Due to overall low prevalence of TGEV, surveillance is very limited. Perhaps federal labs should resume doing surveillance for multiple CoVs important in pigs (PEDV, TGEV) by serology on meat juice samples.
-
Diagnostic kits validated by International, European or National Standards
No. CoVs in pigs are not associated with OIE listed diseases.
-
Diagnostic method(s) described by International, European or National standards
Not applicable.
-
Commercial potential for diagnostic kits worldwide
Depends on the cost and availability. CoV vaccines are not widely used in Europe so there is no immediate need to check for vaccination efficacy using commercial diagnostic kits.
GAPS :
- The biggest issue with multiplex serology is the need for specialized equipment and the great variety/high cost of such equipment. To reduce cost in the long-term and to be able to compare results most labs should utilize the same equipment and share reagents (such as antigen coated beads etc).
- Improved cost-efficient antibody assays, especially multiplex assays would be good to have for routine surveillance which currently is limited to PEDV and TGEV in some countries.
-
DIVA tests required and/or available
There are currently no pig CoV DIVA vaccines. In theory, PRCV infection can be differentiated from TGEV infection/vaccination by ELISA but there is currently no way to differentiate TGEV vaccinated pigs from TGEV infected pigs. The situation is similar for PEDV: Vaccines that could be used are most based on G2b strains which is the most common field strain. The immune response could not be differentiated.Pig CoV vaccines presently do not contain markers.
GAP :
Inserting a foreign marker that could aid as indication of successful vaccination in newly developed CoV vaccines would be an improvement. This would likely need to be driven by pig producers or pig veterinarians. Currently the CoV situation in pigs is stable in Europe and CoV vaccines in pigs are rarely used.
-
Opportunities for new developments
NA.
-
Vaccines availability
-
Commercial vaccines availability (globally)
PEDV vaccines
- Inactivated vaccines
- Region: North America (genotype 2), Asia (genotype 1 or 2 or both)
- Administration: IM to pregnant sows.
- Recombinant alpha virus-based vaccine
- Region: North America (genotype 2),
- Administration: IM to pregnant sows.
- Live attenuated vaccines (monovalent, bivalent or triavalent)
- Region: Asia (China, Japan, South Korea, Philippines)
- Administration: Pregnant sows, IM or orally.
TGEV vaccines
- Live attenuated vaccines (mono, bi- and trivalent for PEDV, rotavirus and/or Escherichia coli.
- Region: North America, Asia.
- Administration: Orally to pregnant sows. The vaccines are often bi- or trivalent vaccines combined with rotavirus, PEDV and/or Escherichia coli.
- Inactivated vaccines (mono, bi- and trivalent for TGEV, rota, PEDV and/or E.coli).
- Region: Asia.
- Administration: IM to pregnant sows.
Bivalent TGEV and PEDV vaccine
- Inactivated vaccines
- Region: Asia (China; strain CV777); Live attenuated tri-valent TGEV, PEDV and porcine rotavirus (China; PEDV strain CV777); Live attenuated vaccine (Japan; PEDV strain 83P-5; South Korea; PEDV strains SM98-1 and DR-13, Philippines, PEDV strain DR13); Inactivated vaccine (South Korea, PEDV strain SM98-1)
Vaccines for CoV have been reviewed in: Gerdts and Zakhartchouk, 2017.
GAPS :
- There are presently no commercial vaccines for PDCoV (low to largely unknown prevalence in Europe), PRCV (only causing subclinical disease, widespread in pigs and thought of as natural protection against TGEV), SeACoV (only found in Asia, possible low prevalence) and PHEV (considered widespread, rare clinical disease outbreaks).
- TGEV is disappearing (not very prevalent) and less commercial vaccines are currently available. The number likely will go further down in future.
- For PEDV few vaccines all containing similar strains are available. Cross-protection among genogroups 1 and 2 is suspected to be limited and PEDV continues to evolve, potentially giving rise to new, more pathogenic strains. It is important to continue to survey novel PEDV variants that may emerge locally or globally through antigenic drift (point mutations) or antigenic shift (recombination events).
- Vaccine platforms that allow a rapid change of strain should be promoted as a precaution.
- Inactivated vaccines
-
Commercial vaccines authorised in Europe
TGEV vaccines have been authorized in selected countries in the past but not used today.See also Section “Commercial vaccines availability (globally)”.
-
Marker vaccines available worldwide
Not available.
-
Marker vaccines authorised in Europe
Not available.
-
Effectiveness of vaccines / Main shortcomings of current vaccines
Cross-protection against different strains/genotypes of the same virus variant (PEDV) has been indicated in experimental settings (Annamalai et al, 2017).Dam vaccination is commonly used to protect the most valuable population, suckling piglets. Protection of piglets depends on successful transfer of immunity to the colostrum and its uptake by the piglet.
GAPS :
- Better ways to protect suckling pigs efficiently in emergency situations i.e antibody supply via other sources (egg antibody etc) that need to be cost efficient and easy to do.
- Possible effective and cost efficient passive vaccination for piglets.
- Improve IgA production in sows without causing disease (good IgA production with MLV vaccines).
-
Commercial potential for vaccines
Depends on disease evolution and effective prevention of incoming new virus variants.
GAPS :
- Vaccine manufacturers all focus on high impact diseases with immediate high sales potential
- Vaccine manufacturer may not be willing to invest into new products when there is no or only a small perceived market.
-
Regulatory and/or policy challenges to approval
GAP :
Need for rapid approval if new/potential strains with higher pathogenic potential are being discovered.
-
Commercial feasibility (e.g manufacturing)
Adequate.
-
Opportunity for barrier protection
Yes.
-
Opportunity for new developments
Several new vaccine platforms have been successfully assessed under experimental conditions and some are available in certain geographic regions outside Europe (Gerdts and Zakhartchouk, 2017).
Examples include:
- Viral vectored vaccines using unrelated viral genomes such as Poxvirus (Hain et al., 2016) or Adenovirus (Liu et al., 2019; Callebaut et al., 1996) to express the CoV spike protein.
- Subunit vaccines in which the spike protein is expressed in mammalian, baculovirus, yeast or plant cells
- Most CoV in pigs affect the intestinal tract. Vaccines directly added in the feed (expressed in Tabaco plants or corn; Lamphear et al., 2004) would be beneficial and make the vaccination process easier.
- DNA vaccines for which genes encoding antigens are cloned into plasmid expression vectors such as Lactobacillus (Liu et al., 2011) or Salmonella (Zhang et al., 2016).
GAPS :
- All of the experimental vaccines described today have their own disadvantages which need to be overcome first to make production successful. Advantages include cost, adjuvant needs, pre-existing immunity against vector, altered immune responses etc.
- Plant expressed vaccines are considered transgenic and would be difficult to get approved in Europe.
-
Pharmaceutical availability
-
Current therapy (curative and preventive)
No curatives (antivirals) are currently available in Europe.
In Asia and North America, chicken egg yolk antibodies have been developed and utilized in pigs to protect pigs in ongoing PEDV outbreaks (especially naïve herds). Specifically, IgY against PEDV S1 protein was used to vaccinate hens. Oral administration of yolks of these hens resulted in protection of neonatal pigs against PEDV (Lee et al., 2015). This method has attracted considerable attention as an alternative to antibiotics to maintain swine health and performance. Oral administration of IgY possesses many advantages over mammalian IgG such as cost-effectiveness, convenience and high yield (Li et al., 2015).
GAPS :
- Only useful when inexpensive in production.
- Application by drinking water or via feed is preferred but injectable may be acceptable for pigs.
- Withdrawal period should be very short.
-
Future therapy
Very unlikely.
GAP :
Coronaviruses have a potential to evolve quickly. Studies would be required to assess the potential of escape mutants to the drug.
-
Commercial potential for pharmaceuticals
Very low.
GAP :
Retailer and authority approval needed before it could be used.
-
Regulatory and/or policy challenges to approval
Only useful when inexpensive, application by water or feed preferred but application via injection would be acceptable. Withdrawal period should be very short.
-
Commercial feasibility (e.g manufacturing)
See section ‘Regulatory &/or policy challenges to approval’.
GAP :
Treatment should be inexpensive (cents to one euro per pig) to be attractive.
-
Opportunities for new developments
See section ‘Regulatory &/or policy challenges to approval’.
-
New developments for diagnostic tests
-
Requirements for diagnostics development
Any new virus detection (RNA, antigen) test needs to be accurate, inexpensive, fast (24 hours or less), and not require expensive, sensitive equipment. As most PigCoV target the enteric tract the ideal sample type is faeces or rectal swabs.
There may also be a need for advanced immunity determination of a pregnant sow. At approximately 3 weeks prior to farrowing: Can it be predicted that she will transfer sufficient immunity in colostrum/milk. If not, the dam could be re-vaccinated at that time.
-
Time to develop new or improved diagnostics
Unknown, depends on diagnostic assay targeted and diagnostic platform.
-
Cost of developing new or improved diagnostics and their validation
Variable depending on platform used and the need to create samples for test validation (or just purchase positive and negative controls).
-
Research requirements for new or improved diagnostics
- Pen-side tests using fecal samples, rectal swabs, blood, or colostrum collected at the site would be beneficial.
- Sequencing to determine mutations needs to be much faster. In other regions outside Europe this may be done in 24 hours or less while in Europe this often requires shipment to specialized labs and potential long waiting times.
GAPS :
- Comparisons among laboratories are needed but rarely done on a regular basis.
- Control samples from different geographic areas should be shared among labs or offered for purchase by an independent source.
-
Technology to determine virus freedom in animals
Improved tests with high sensitivity but also capable to detect mutants (broader assays).
GAPS :
- Tests may often have high sensitivity but virus mutants or other CoV species may not be detected readily
- It would be useful to use two tests on a regular basis: One for genetic CoV and one for the desired virus i.e. PEDV, TGEV etc.
-
New developments for vaccines
-
Requirements for vaccines development / main characteristics for improved vaccines
Multiple strain coverage (multivalent, several CoV and several types of each CoV), easy to apply (mass vaccination), single dose, inexpensive vaccines, good transfer from sow to the piglet.
-
Time to develop new or improved vaccines
Variable depending on desired vaccine type.
-
Cost of developing new or improved vaccines and their validation
Variable, depends on vaccine platform utilized and technical expertise.
-
Research requirements for new or improved vaccines
- Development of recombinant vaccines, subunit vaccines and possibly DNA vaccines.
- Research requirements would change based on targeted platform.
GAP :
Costs relating to the research and development of some types of vaccines in the face of a potentially limited market.
-
New developments for pharmaceuticals
-
Requirements for pharmaceuticals development
Unknown.
-
Time to develop new or improved pharmaceuticals
Unknown, many years before it could be on the market.
-
Cost of developing new or improved pharmaceuticals and their validation
Unknown, very high.
-
Research requirements for new or improved pharmaceuticals
Unknown, however strict assessment of the potential for new pharmaceuticals to generate escape mutants should be a priority.
Disease details
-
Description and characteristics
-
Pathogen
The family Coronaviridae within the order Nidovirales consists of two subfamilies: (1) Orthocoronavirinae comprising the genera Alphacoronavirus, Betacoronavirus, Gammacoronavirus,and Deltacoronavirus and (2) Letovirinae comprising the genera Alphaletovirus.
There are currently six known coronaviruses (CoVs) circulating in the global pig population (Table 1) (Saif et al., 2019):
- TGEV first described in 1946
- PRCV, a spike (S) gene deletion mutant of TGEV isolated in 1984
- PEDV isolated in 1977
- pHEV isolated in 1962
- PDCoV detected in 2012
- SeACoV, a bat‐HKU2‐like alphacoronavirus has been identified in swine in China in 2017 (Gong et al., 2017; Pan et al., 2017).
These are classified as follows according to the ICTV (2020):
Order: NidoviralesSuborder CornidovirineaeFamily: CoronaviridaeSubfamily: OrthocoronavirinaeGenus: Alphacoronavirus, Betacoronavirus, and Deltacoronavirus
A TGEV/PEDV recombinant virus (TGEV backbone but with PEDV spike gene) has been identified in swine in Europe in 2016 (Akimkin et al., 2016; Belsham et al., 2016; Boniotti et al., 2016).
CoVs are enveloped and pleomorphic with an overall diameter of 60-120nm. Most CoVs contain four structural proteins: a large surface glycoprotein (spike or S protein visible as the corona), a small membrane protein (E), an integral membrane glycoprotein (M), and a nucleocapsid protein (N). However, pHEV also contains an HE protein (Saif et al., 2019).
Table 1. Described coronaviruses in pigs (Saif et al., 2019)
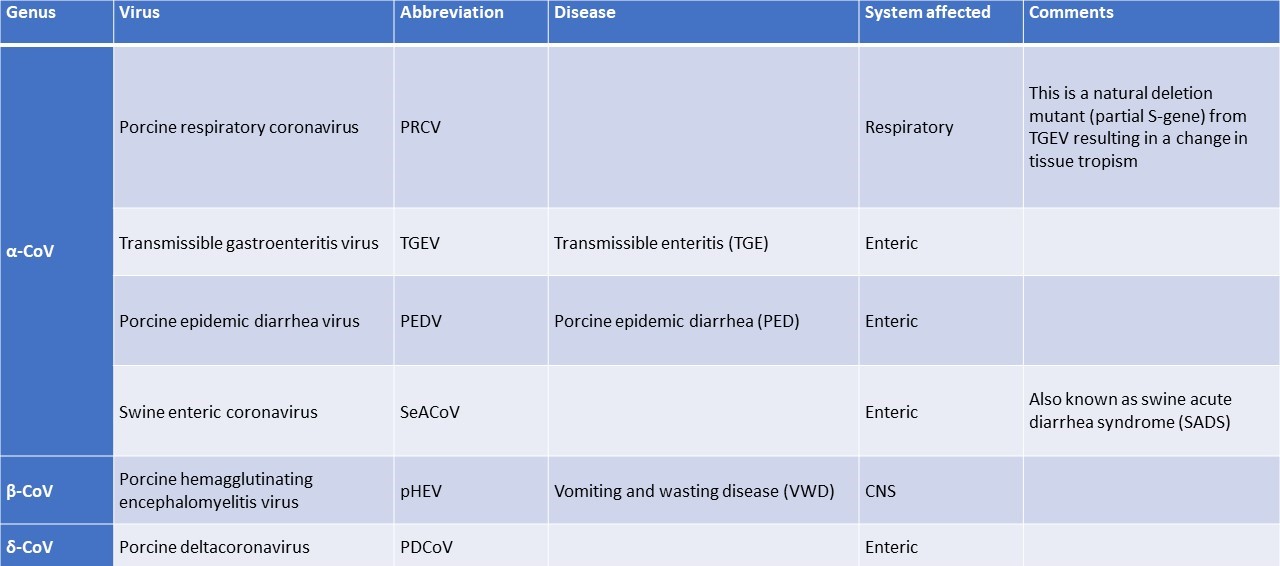
-
Variability of the disease
PRCV: PRCV isolates generally fall into two categories, European or U.S., though other isolates have been described in Canada (Jabrane and Elazhary, 1994), Japan (Usami et al., 2008), and Korea. The European isolate arose in Belgium (Pensaert et al., 1986) and then spread throughout Europe, whereas the U.S. isolates (at least 7 have been described) appear to have arisen independently (Weley et al., 1990). Variability between strains occurs in the size of the deletion within the S gene (600-700 base pairs) (Usamie et al., 2008; Vaughn et al., 1994) as well as one of the subgenomic RNAs, open reading frame 3 and 3–1, which can be found during replication (Zhang et al., 2007). Virulence in pigs varies with the isolate. The disease associated with PRCV is mild to moderate respiratory disease with some strain variation when PRCV first appeared (Vaughn et al., 1994). Today most pigs are thought to be subclinically infected with PRCV.
TGEV: There are two main clusters: Miller and Purdue strains. The complete genomes of the Purdue and Miller strains of TGEV are 28546–28580 nucleotide long and share 96% overall identity (Penzes et al. 2001; Zhang et al. 2007). Natural recombinant strains of TGEV between the Purdue and Miller clusters were isolated from the small intestine content of piglets in China (Zhang et al., 2017). There is no major clinical difference among strains.
PEDV: Two genogroups have been described, G1 (containing classical strains) and G2 (containing field epidemic or pandemic strains). Genogroup 1 can be further subdivided into G1a which contains the vaccine strain CV777 and other cell culture adapted strains and G1b which contains new PEDV variants present in Asia and the USA which are also known as S-INDEL strains (due to insertion and deletions in the spike gene region). Genogroup 2 can be further divided into G2a which contains strains previously involved in epidemic outbreaks in Asia and G2b which contains recent pandemic strains involved in outbreaks in Asia and North America (Lee, 2015).
SeACoV: No information.
pHEV: Overall the homology among strains appears high. However, not many stains have been sequenced and are not available in Genbank (Li et al., 2016).
PDCoV: All global PDCoV strains share high nucleotide identities (Zhang 2016) with region specific clusters (Zhang et al., 2019).
GAP :
To properly assess disease variability among PigCoV sub-strains regular infection studies under controlled similar conditions would need to be conducted.
-
Stability of the agent/pathogen in the environment
PRCV: Survival of PRCV in the environment is unclear (Killoran et al., 2016). In PRCV endemic herds, the virus can be isolated from pigs throughout the year. In other herds, PRCV temporarily disappears during summer months. PRCV may be highly stable when frozen, as is TGEV (Killoran et al., 2016).
TGEV: TGEV is stable when stored frozen, but labile at room temperature or higher. Infectious virus persisted in liquid manure slurry for more than 8 weeks at 5°C, 2 weeks at 20°C, and 24 hours at 35°C (Haas et al. 1995). In recent studies that used TGEV as a surrogate for SARS CoV (Casanova et al. 2009), it remained infectious in water and sewage for several days at 25°C and for several weeks at 4°C. TGEV is highly photosensitive. Fecal material containing 1 × 105 pig infectious doses (PID) was inactivated within 6 hours when exposed to sunlight or to ultraviolet light (Cartwright et al. 1965).TGEV is inactivated by exposure to 0.03% formalin, 1%Lysovet (phenol and aldehyde), 0.01% beta‐propiolactone, 1 mM binary ethylenimine, sodium hypochlorite, NaOH, iodines, quaternary ammonium compounds, ether, and chloroform (Brown 1981; VanCott et al. 1993). TGEV field strains are trypsin resistant, relatively stable in pig bile, and stable at pH 3 (Laude et al. 1981), allowing the virus to survive in the stomach and small intestine. However, properties of attenuated and field strains of TGEV vary.
PEDV: The virus can survive for variable periods outside the host depending on the temperature and relative humidity, for example, it can survive at least 28 days in slurry at 4°C, 7 days in faeces-contaminated dry feed at 25°C, up to 14 days at 25°C in wet feed and at least 28 days in wet feed mixture at 25°C. The virus loses infectivity above 60 °C. PEDV is susceptible to Formalin (1%), Anhydrous sodium carbonate (4%), lipid solvents, iodophores in phosphoric acid (1%), Sodium hydroxide (2%). It is stable at pH 6.5-7.5 at 37°C and pH 5-9 at 4°C (OIE Technical Fact Sheet).
SeACoV: No information.
pHEV: Exposure of pHEV to 37°C results in loss of infectivity over a period of three days (Paensert and Callebaut, 1974). PHEV, like other CoVs, is highly stable when frozen and at low temperatures (Alsop, 2006). In winter, PHEV can survive for extended periods of time. Exposure to ultraviolet light for two minutes inactivates PHEV (Greig and Bouillant, 1972, Killoran et al., 2018).PHEV is relatively stable at pH 3.0, losing only 20% infectivity after 24 hours (Paensert and Callebaut, 1974). The virus may also lose infectivity at alkaline pH values, as do other CoVs (Saif et al., 2019).1.
PDCoV: No information available.
GAP :
It appears that stability studies are commonly done with single strains of a CoV. It seems necessary to compare strain and viruses at the same time.
-
Species involved
-
Animal infected/carrier/disease
Pigs are the main species naturally susceptible to the described pig CoVs. No infection of humans has been reported (Saif et al., 2019).
-
Human infected/disease
None reported to date.
-
Vector cyclical/non-cyclical
None identified.
-
Reservoir (animal, environment)
PRCV: Pig farms where the virus is present in subclinically infected pigs. This also includes wild boars.
TGEV: Pig farms where the virus is present in subclinically infected pigs. Non-porcine animals such as cats, dogs, and foxes have been suggested as possible carriers of TGEV from one herd to another, since they can shed the virus in their feces for variable periods (Haelterman 1962; McClurkin et al. 1970) and virus excreted by dogs was infectious for pigs (Haelterman 1962; Reynolds and Garwes 1979). The concentration of starlings (Sturnus vulgaris) in winter in feeding areas of swine may increase the mechanical spread of TGEV among farms. TGEV has been detected in the droppings of starlings for up to 32 hours after feeding TGEV (Pilchard, 1965). Houseflies (Musca domestica) have also been proposed as possible mechanical vectors for TGEV. TGEV antigen was detected in flies within a swine herd, and experimentally inoculated flies excreted TGEV for 3 days (Gough and Jorgenson 1983). According to surveys conducted in Central Europe, antibodies against TGEV are also present in approximately 30% of the feral pig population (Sedlak et al. 2008).
PEDV: Pigs and wild boars (Lee et al., 2016) are the only reservoir for PEDV.
SeACoV: No information on non-pig reservoirs are available at this point.
pHEV: Pigs are the only species naturally susceptible to pHEV. No other reservoirs have been demonstrated, although the virus can be experimentally adapted to mice and Wistar rats (Killoran et al., 2018).
PDCoV: Pigs are the only known reservoir for PDCoV. Deltacoronaviruses also occur in birds but are different from PDCoV (Ma et al., 2015).
GAP :
With the better ability of research reagents including secondary antibodies for various species, possible non-pig reservoirs for known pig CoVs should be further investigated.
-
Description of infection & disease in natural hosts
-
Transmissibility
PRCV:PRCV infects pigs of all ages by contact or airborne transmission. PRCV infections are often subclinical (Saif et al., 2019). The risk of PRCV spread increases in areas of high swine density, where the virus can travel several kilometers. The virus has spread rapidly and extensively in pigs in Europe (Brown and Cartwright 1986; Have 1990; Laude et al. 1993) and became endemic even in TGEV‐free countries (Laude et al. 1993, Pensaert et al. 1993). A limited serological survey in 1995 in the United States suggested that many asymptomatic herds in Iowa were seropositive for PRCV (Wesley et al. 1997). PRCV spread usually occurs post-weaning when maternally derived antibody-mediated protection begins to decline. Transmission may also occur in growers/finishers when PRCV-naïve pigs are introduced (Killoran et al., 2016).
TGEV: Although feces are the major source of infection, the virus is probably spread aerogenously, at least for short distances. In feeding operations or multiple farrowing operations, where animals from multiple sources are intermixed, carrier animals often are a source of exposure to TGE virus. Other animals and insects known to act as mechanical carriers of virus for various lengths of time and distance include dogs, cats, foxes, starlings and flies. The virus is resistant enough that fomites readily transmit the virus. Once introduced, the virus may persist on premises, especially during the colder months. Conversely, premises depopulated for a few days during hot summer months may then be free of virus (TGE ISU Swine Manual).
PEDV: Direct transmission occurs through ingestion of virus-contaminated faeces (OIE PEDV Fact Sheet). Indirect transmission occurs through vehicles which may be contaminated including feed trucks, service vehicles as well as personnel, equipment or other types of faeces-contaminated objects including feed. Contaminated pig blood products, such as spray-dried plasma, that are incorporated into rations for feeding piglets have been suspected as a possible means to spread the virus. However, multiple experimental studies suggested that spray-dried porcine plasma is not a likely source of infectious virus provided that good manufacturing practices and biosecurity standards are followed. Contaminated vehicles used for the movement of pigs have been identified as an important risk factor for spreading the disease (OIE PEDV Fact Sheet).
SeACoV: Oral-faecal transmission (Yang et al., 2020).
pHEV: The virus is transmitted by direct nose-to-nose contact, aerosols, and contaminated fomites (Killoran et al., 2018).
PDCoV: The main transmission is via the faecal–oral route. After experimental infection pigs shed the virus for up to 19 days in the faeces. Faeces, vomit and other contaminated fomites are the major sources of the virus (Vlasova et.al., 2020).
-
Pathogenic life cycle stages
Not applicable.
-
Signs/Morbidity
Enteric coronaviruses (TGEV, PEDV, SeACoV, PDCoV,) cause very similar clinical signs and cannot be distinguished without virus/RNA detection.
PRCV (Killoran et al., 2016)
- Commonly subclinical
- Pigs can display polypnea, dyspnea, tachypnea, sneezing, coughing, fever, anorexia, and delayed growth.
- Severe respiratory stress is rarely seen.
- In experiment infected pigs, clinical signs appear between 4-10 DPI and resolve by 14 DPI.
TGEV (TGE ISU Swine Manual)Acute outbreaks:
- Rapid spread in baby pigs; large number of affected piglets
- Profuse diarrhea
- Frequent vomiting.
- Rapid dehydration, shivering and marked thirst
- Piglets weaken rapidly and usually die within 1-2 days.
- Pigs suckling immune dams may remain well as long as they receive adequate antibody in the dam’s colostrum and milk.
- Pigs infected after 4 weeks of age often survive.
Chronic or endemic form of TGE:
- Signs can be rather mild, especially in well-doing pigs, and usually include diarrhoea, dehydration, thriftiness and runting.
Feeder and fattening pigs:
- Signs are usually mild except for diarrhea which is profuse and watery for a few days.
- Vomiting occurs occasionally.
- Morbidity is high but mortality is low or absent.
- Moderate severity is observed in naïve sows and gilts, especially in those that have farrowed recently and are heavily exposed to virus from piglets with TGE.
- The dams show anorexia, vomiting, diarrhea, depression and may cease to lactate. Recovery usually occurs within 5-10 days.
PEDV (OIE PEDV fact sheet):
- Morbidity: up to 100%,
- Mortality varying according to age:
- Suckling piglets: up to 100%
- Piglets older than 10 days: less than 10%
- Adult and fattening pigs: less than 5%
- Diarrhoea and vomiting.
- Dehydration and metabolic acidosis.
SeACoV: Variable based on few challenge studies (Yang et al., 2020).
- Diarrhea
- Vomiting
- Dehydration
- No clinical signs
pHEV (Saif et al., 2019)
- Sneezing or coughing with elevated body temperature for up to two days.
- 2 main clinical forms in pigs below 3-4 weeks of age:
- VWD (Vomiting and wasting disease): frequent vomiting leading to death or subsequent wasting. Pigs start suckling from the sow but stop, withdraw from the sow and vomit the milk they have consumed.
- Acute encephalomyelitis with motor disorders such as generalized muscle tremor, hyperaesthesia, inability to stand, jerky gait, tend to walk backwards, blindness.
PDCoV (Saif et al., 2019)
- Clinical signs in suckling and older pigs are similar, but milder, than those of PEDV and TGEV infections.
- Acute, watery diarrhea, frequently accompanied by vomiting, leading to dehydration, loss of body weight, lethargy and death.
GAP :
Side by side experimental pig infections may be useful to further clarify if there are clinical differences or differences in viral shedding and immune response among pig CoVs.
-
Incubation period
PRCV: 2-4 days (Killoran et al., 2016)
TGEV: 18 hours to 3 days (TGE ISU Swine Manual)
PEDV: 1-4 days (OIE PEDV Fact Sheet)
SeACoV: Short but no detailed information (Yang et al., 2020).
pHEV: 4-7 days (Saif et al., 2019).
PDCoV: 1-3 days (Saif et al., 2019).
-
Mortality
PRCV: Very low to none.
TGEV: Minimal to high depending on age of infection (high in very young pigs) and status of the farm (acute or chronic infected).
PEDV: The virus will cause clinical signs in swine of all ages. Older animals will recover. Mortality in piglets (< 14 days of age) from a naïve herd will range from 30-100%. This virus appears to be particularly pathogenic with losses in young pigs approaching 100% in most cases.
SeACoV: Variable information based on few studies ranging from 0-100% (Yang et al., 2020).
pHEV: 100% in neonatal pigs and mild transient illness in older pigs . Outbreaks described in Taiwan (Chang et al. 1993) in 30‐to 50‐day‐old pigs indicated a morbidity of 4% and a mortality approaching 100%. The pigs died 4–5 days after the onset of clinical signs.
PDCoV: Up to 40% (USA) to 64-80% (Asia) mortality has been observed in suckling pigs.
GAPS :
- Side by side controlled infection studies are needed to assess if there are major differences in mortality in pigs infected with different CoVs.
- More work is needed to assess the potential importance of SeACoV.
-
Shedding kinetic patterns
PRCV: 2-10 days in nasal swabsNasal shedding of PRCV in experimentally infected pigs occurs through 10 days post infection (DPI) (Onno et al. 1989; Wesley et al. 1990). In another study virus was recovered from experimentally infected pigs in air samples for 6 days after infection (Bourgueil et al., 1992). The highest quantities of virus were recovered on days 2 and 4 post-infection. Concurrently collected nasal swab samples were positive for PRCV for 8 days post-infection (DPI) (Bourgueil et al.,1992).
TGEV: Chronic and/or persistent TGEV fecal shedding has been reported for up to 18 months, suggesting a possible role for the long‐term carrier hog in transmitting TGEV (Woods and Wesley 1998). Although TGEV has been detected in the intestinal and respiratory tracts for periods of up to 104 DPI (Underdahl et al. 1975), it is unknown whether infectious virus is shed or transmitted. Addition of sentinel pigs to a herd at 3, 4, and 5 months after a previous TGE outbreak resulted in no infections in the introduced pigs, as determined by serologic tests (Derbyshire et al. 1969).
PEDV: RNA shedding in faeces or rectal swabs can be detected from 1 or 2 days until approximately 21 DPI with a peak between 5-6 DPI. However, PEDV RNA can be detected in individual pigs at least for 28 DPI (Niederwerder et al., 2016).
SeACoV: There are only few studies and mixed results. Shedding is not described in detail.
pHEV: When 7 week old pigs were infected, viral RNA could be demonstrated in faeces between 1-10 DPI (Mora-Díaz et al., 2019).
PDCoV: Diarrhea in infected piglets was observed for approximately 5–10 days, with persisting viral RNA shedding for up to 19–28 days in feces and for up to 42 days in oral fluids (Saif et al., 2019).
-
Mechanism of pathogenicity
PRCV: PRCV replicates in the upper and lower respiratory tract (alveolar cells, nasal mucosa, tracheal, bronchial,and bronchiolar epithelium, alveolar macrophage, and tonsils) and can be isolated readily from nasal swabs for 6–10 days post-infection (Killoran et al., 2016). Replication in the intestine has been shown by some to occur in a few, unidentified cells located underneath the epithelial layer (Killoran et al., 2016).
TGEV: Jejunal enterocytes undergo massive necrosis within 12–24 hours after infection, resulting in marked reduction in enzymatic activity (alkaline phosphatase, lactase, etc.) in the small intestine (Saif et al., 2019). This disrupts digestion and cellular transport of nutrients and electrolytes (including sodium), thereby causing an accumulation of liquid in the intestinal lumen and acute malabsorptive diarrhea (Moon 1978) that leads to severe and fatal dehydration in piglets (Butler et al. 1974) and loss of extravascular protein. Dehydration is also related to metabolic acidosis coupled with abnormal cardiac function due to hyperkalemia. The severe villous atrophy in the jejunum and to a lesser extent in the ileum of TGEV infected pigs is often absent in the proximal duodenum (Hooper and Haelterman 1966). Villous atrophy is more severe in newborn pigs than in 3‐week‐old pigs (Moon 1978). Extraintestinal sites for TGEV replication include lungs (alveolar macrophages) and mammary tissues (Kemeny et al. 1975). Oronasal infection of pigs with TGEV caused pneumonia (Underdahl et al. 1975). TGEV replicated in mammary tissues of lactating sows (Saif and Bohl 1983) and infected sows shed virus in milk (Kemeny and Woods 1977).
PEDV: Oral ingestion results in viral replication in the epithelial cells of the small intestinal and colonic villi resulting in degeneration of enterocytes leading to shortening of the villi (OIE PEDV Fact Sheet). This causes clinical manifestations of the disease including watery diarrhea.
SeACoV: Unknown, not further described yet.
pHEV: The virus first replicates in the nasal mucosa, tonsils, lungs, and to a very limited extent, in the small intestine (Pensaert, 2013). From these sites of entry, the virus invades defined nuclei of the medulla oblongata via the peripheral nervous system and subsequently spreads to the entire brain stem, and possibly to the cerebrum and cerebellum. Vomiting is thought to be caused by viral replication in the vagal sensory ganglion. Wasting is due to vomiting and delayed emptying of the stomach, which is the result of virus-induced lesions in the intramural plexus. Infection of cerebral and cerebellar neurons may cause motor disorders.
PDCoV: Replication of PDCoV is confined to the small and large intestinal epithelia. PDCoV-infected enterocytes rapidly undergo acute necrosis, leading to marked villous atrophy in the small intestine but not in the large intestine (Saif et al., 2019). During acute infection, PDCoV antigens are detected mainly in the villous epithelium of the atrophied mid‐jejunum to ileum and, to a lesser extent, in duodenum, proximal jejunum, and cecum/colon. Frequently, acute viremia with low PDCoV RNA titers in serum was observed (Saif et al., 2019).
-
Zoonotic potential
-
Reported incidence in humans
None.
-
Risk of occurence in humans, populations at risk, specific risk factors
Very low to non-existent.
-
Symptoms described in humans
Very low to non-existent.
-
Estimated level of under-reporting in humans
None described.
-
Likelihood of spread in humans
Not applicable.
-
Impact on animal welfare and biodiversity
-
Both disease and prevention/control measures related
Depends on the virus involved. Severe impact on animal welfare when naïve herds become infected with PEDV or TGEV.
-
Endangered wild species affected or not (estimation for Europe / worldwide)
Pigs are the only known host and this also includes wild boars. There are no endangered species affected by the virus.
-
Slaughter necessity according to EU rules or other regions
Slaughter is not necessary according to EU rules.
-
Geographical distribution and spread
-
Current occurence/distribution
TGEV: Europe, Asia, the Americas and AfricaPRCV: WorldwidePEDV: Classical and emerging non-S INDEL and S-INDEL strains Asia and Europe. Americas uniquely emerging non-S INDEL and S-INDEL strains.PDCoV: Asia and North AmericapHEV: World wideSeACoV: China(Vlasova et al., 2020; Wang et al., 2019).
-
Epizootic/endemic- if epidemic frequency of outbreaks
TGEV: Endemic and epidemic (sporadic cases)PRCV: Endemic worldwidePEDV: Continual epidemic outbreaks Europe, China and the Americas since the 1970’s. Most recent outbreaks mainly concern non-S INDEL and S-INDEL strains. Can become endemic if the virus is maintained due to continued production and weaning in large numbers.PDCoV: Newly emerging pig coronavirus. Has been reported at high prevalence and frequently in co-infections with PEDV (Dong et al., 2015; Song et al., 2015) pHEV: Endemic, but most cases subclinical.SeACoV: China only, frequency unknown for the moment.
-
Seasonality
At colder temperatures, coronaviruses in general have increased survival times outside of the host, thus infections will be generally more frequent in the winter months.
-
Speed of spatial spread during an outbreak
These viruses are in general highly infectious and thus the speed of spread will depend largely on environmental conditions, level of hygiene, biosecurity, density of animals efficacy of vaccination (where applicable see also section “Vaccines availability”) and vaccination programs on farms on and around an outbreak.
-
Transboundary potential of the disease
Occurrence of pig CoVs is often transboundary.
-
Route of Transmission
-
Usual mode of transmission (introduction, means of spread)
TGEV, PEDV, PDCoV and SeACoV: in line with other enteric coronaviruses the usual mode of transmission is faecal-oral via direct or indirect methods.PRCV and pHEV: Direct contact or aerosols.
-
Occasional mode of transmission
Mechanical: contaminated food stuffs, farm workers clothing, vehicles and other fomites.
-
Conditions that favour spread
High density farming, poor hygiene measures.
-
Detection and Immune response to infection
-
Mechanism of host response
PRCV: Serum neutralizing antibodies can be detected beginning around 6 DPI upon primary infection with PRCV (Killoran et al., 2016). The antibody response peaks approximately 14 DPI and subsequently wanes.Following PRCV infection, neutralizing antibodies can be found in milk at minimal levels from 7–14 days post-farrowing with an increase in titers as lactation continues. Milk IgA levels vary among individuals following a single infection and reinfection with PRCV results in an increase in IgA detected in milk. The duration of effective PRCV-induced immunity appears to be relatively short lived. PRCV-induced neutralizing antibody (nAb) titers are high at three weeks post-infection (WPI), low by 36 WPI , and marginal to absent one year post-infection. Repeated infection with PRCV can be achieved. Additionally, within 1–2 weeks post-weaning, pigs become susceptible to PRCV infection as passive immunity wanes (Killoran et al., 2016).An immune response to PRCV is mainly relevant to prevent TGEV infection. A partial immunity against TGEV in PRCV infected pigs may be related to the rapid increase in TGEV VN antibodies (Cox et al. 1993; Wesley and Woods 1996) and numbers of IgG and IgA antibody secreting cells (ASCs) in the intestines of PRCV‐exposed pigs after TGEV challenge (Saif et al. 1994; VanCott et al. 1994).
TGEV: Serum antibodies provide serological evidence of TGEV or PRCV infection, they afford little indication of the degree of active immunity to TGEV (Saif et al., 2019).Swine that have recovered from TGE are immune to subsequent short‐term challenge, presumably due to local immunity within the intestinal mucosa (Brim et al. 1995; Saif et al. 1994; VanCott et al. 1993, 1994). The age and immune status of the animal at initial infection and the severity of the challenge influence the completeness and duration of active immunity. The mechanism of active immunity in the gut relates to stimulation of the secretory IgA (sIgA) immune system with production of sIgA antibodies by intestinal plasma cells (Saif et al. 1994; VanCott et al. 1993, 1994). IgA TGEV antibodies and ASCs have been detected in the intestine and serum of pigs after oral, but not parenteral inoculation with TGEV (Kodama et al. 1980; Saif et al. 1994; VanCott et al. 1993, 1994). Kodama et al. (1980) proposed that detection of IgA antibody in the serum, presumably intestinally derived, might serve as an indicator of active immunity to TGEV. Enzyme‐linked immunospot (ELISPOT) assay was used to investigate the kinetics of IgA and IgG TGEV antibody production by the pig’s systemic and local gut-associated lymphoid tissues (GALT). High numbers of IgA ASCs were induced in GALT only by virulent TGEV. CMI was demonstrated with lymphocytes obtained from GALT of swine orally infected with virulent TGEV (Brim et al. 1995; Frederick et al. 1976; Shimizu and Shimizu 1979), whereas swine parenterally or oronasally inoculated with attenuated TGEV or PRCV developed CMI mainly in systemic sites. Although PED occurs in pigs of all ages, piglets up to 1 week of age may experience high mortality and need to be protected by maternal antibodies, especially VN and sIgA, via colostrum and milk from immunized dams (Saif et al., 2019).
PEDV: Humoral immunity to PEDV is very similar to that induced by TGEV. However, there is evidence that PEDV has the ability to evade host IFN responses. Of 21 PEDV‐encoded proteins, at least 11 proteins have been identified as IFN antagonists, which include both ORF1ab‐encoded NS proteins (nsp1, nsp3, nsp5, nsp7, nsp14, nsp15, nsp16), structural proteins (E, M, N), and the accessory protein ORF3 (Ding et al. 2014; Wang et al. 2015; Zhang et al. 2016).
SeACoV: Not investigated at this point.
pHEV: After infection, pigs develop detectable protective circulating antibodies (hemagglutination inhibition assay, virus neutralization assays) to pHEV in 7–9 days (Saif et al., 2019). PHEV-neutralizing antibodies are transferred in colostrum and milk from PHEV-seropositive sows to their offspring. Neutralizing antibodies are first detectable between 6–9 days post-infection, very soon after the development of clinical signs (Killoran et al., 2018). The duration of immunity is less important in pHEV because of the resistance to disease that develops with age. Neonatal pigs born to immune mothers are fully protected by maternally derived antibodies that persist until the age of 4–18 (mean 10.5) weeks (Paul and Mengeling 1984). Passive immunity lasts from 8-18 weeks of age.
PDCoV: The immune responses of pigs to PDCoV infection are largely undefined. Development of PDCoV antibodies in serum of PDCoV‐infected pigs has been described (Hu et al. 2016). In experimentally infected pigs, serum IgG, IgA, and VN antibodies were present by 14 DPI and peaked at 24 DPI, when the pigs had recovered from clinical disease and fecal virus shedding stopped (Hu et al. 2016).
-
Immunological basis of diagnosis
PRCV: IgG antibodies in serum (no good differentiation between PRCV and TGEV infection).
TGEV: IgG or antibodies in serum (no good differentiation between PRCV and TGEV infection).
PEDV: IgG or IgA antibodies in serum or faecal sample.
SeACoV: Unknown, immunology has not been described yet.
pHEV: Protective circulating antibodies can be detected by hemagglutination inhibition or virus neutralization assays on serum (Saif et al., 2019). Recently an ELISA based on the pHEV spike gene has also been validated and could be used to detect IgG antibodies in piglets or dams (Mora-Díaz et al., 2020). In addition, an immunochromatographic strip was developed for the detection of pHEV antibodies using colloidal gold-labelled rabbit anti-pig immunoglobulin G (Chen et al., 2011). This strip has the potential to be used on site (Chen et al., 2012).
PDCoV: IgG or IgA antibodies in serum or faecal samples.
GAP :
Cellular immunity is almost never determined although considered important. Current tests done by few specialized labs require that blood samples arrive in the lab within 2 hours after collection. Easier tests or methods to preserve blood on farm prior to testing to determine cell-mediated immunity need to be developed.
-
Main means of prevention, detection and control
-
Sanitary measures
CoV’s are enveloped viruses and therefore sensitive to treatment with soaps and disinfectants.
-
Mechanical and biological control
Cleaning, disinfection and biosecurity.
-
Diagnostic tools
See section “Diagnostics availability”.
-
Vaccines
See section “Vaccines availability”.
-
Therapeutics
None.
-
Biosecurity measures effective as a preventive measure
Strict hygiene, disinfection, reduced number and ensuring adequate hydration are crucial to reduce the severity and duration of infectious episodes. Vaccination is also employed for PEDV (US and Asia) and TGEV although usage of TGEV vaccines is dropping as pigs have natural protection against TGEV following infection with PRCV.
-
Border/trade/movement control sufficient for control
Regulations and controls are in place concerning PEDV (for example in France) and TGEV (for example in Germany).
-
Prevention tools
Biosecurity and vaccination (US and Asia against PEDV).
-
Surveillance
Passive in countries imposing regulations (see section Border/trade/movement control (management) sufficient for control).
-
Past experiences on success (and failures) of prevention, control, eradication in regions outside Europe
Eradication of Pig CoVs like AvCoVs has never been successful, prevalence is also far too high.
-
Costs of above measures
Reasonable too high in countries requiring vaccination programmes.
-
Disease information from the WOAH
-
Disease notifiable to the WOAH
No.
-
WOAH disease card available
OIE technical factsheet on infection with PEDV.
-
WOAH Terrestrial Animal Health Code
-
WOAH Terrestrial Manual
-
Socio-economic impact
-
Zoonosis: impact on affected individuals and/or aggregated DALY figures
Not applicable, circulating CoV in pigs are not zoonotic and do not infect humans.
-
Zoonosis: cost of treatment and control of the disease in humans
Not applicable.
-
Direct impact (a) on production
In general, both PEDV and TGEV are capable of having a major impact on pig production. The actual impact caused depends on the disease status of the farm and region (free/naïve population, acutely infected, chronically infected). The impact of PEDV on a PEDV naïve population has been demonstrated by introduction of PEDV into the North American pig population during 2013 (Stevenson et al., 2013). When comparing pig inventory data for the first 12 months after PEDV introduction (2013 to 2014) the entire U.S pig inventory was reduced by 3.725.000 head (Schulz and Tonsor, 2015). In addition, PEDV introduction also lead to trade issues between North America and PEDV free countries.
The impact of PRCV, SeACoV, pHEV and PDCoV is currently very low and negligible.
-
Direct impact (b) cost of private and public control measures
Little direct impact of control measures: Biosecurity measures need to be in place for various pig diseases and not just pig CoVs including acclimation periods for incoming pigs, all-in, all-out principles, change of clothes and boots, strict disinfection regimens, limit to visitors, disinfection of incoming vehicles.
-
Indirect impact
Indirect impact is mainly a disruption of production. Suckling pigs are mainly affected and hence the upstream production sites cannot be filled.
-
Trade implications
-
Impact on international trade/exports from the EU
Possible concerning regulations in some countries for TGEV and PEDV.
-
Impact on EU intra-community trade
Possible concerning EU regulations in some countries for TGEV and PEDV.
-
Impact on national trade
Possible concerning regulations in some countries for TGEV and PEDV.
-
Main perceived obstacles for effective prevention and control
- Strain variation, development of new variants and hence differences in the sensitivity of diagnostic assays.
- Global travel and trade enhances unintended introduction in areas were certain CoVs are not present.
- Movement of infected pigs may enhance risk of introduction on a farm or in an area.
- Potential trade implications of live pigs from areas infected with some of the CoVs.
- High mortality in piglets in new introductions can be disturbing to pig owner and public.
-
Main perceived facilitators for effective prevention and control
- Food security considerations.
- Production facilities.
- Ability to trade pigs with other countries.
- Early diagnosis (routine surveillance) can result in timely vaccination and lower loss of animals.
- Timely diagnosis to detect the pathogen as quick as possible: reduction of times to the lab and turn-around of the test.
- Cost efficiency of routine testing and vaccination.
GAP :
Early detection and timely notification.
-
Links to climate
Seasonal cycle linked to climate
See section “Seasonality”.
-
Distribution of disease or vector linked to climate
See section “Seasonality”.
-
Outbreaks linked to extreme weather
Not relevant.
-
Sensitivity of disease or vectors to the effects of global climate change (climate/environment/land use)
Unknown.
Risk
-
- Inactivated vaccines or subunit vaccines have little risk.
- Vaccines containing attenuated live virus strains could contribute to new more pathogenic strain evolvements.
- It may be beneficial to determine the virus strain that is present on a farm and use this information to guide vaccine selection if possible and if attenuated live virus strains are used.
Main critical gaps
-
- Real time detection tool of various CoVs that is time sensitive and cost efficient.
- Readily available vaccine platforms based for example on virus/bacteria vector systems where genes of new strains can be inserted rapidly and which can be expanded rapidly if needed.
- As most CoVs target the enteric system, development of oral live vector vaccines that are not composed of whole live CoVs should be promoted.
Conclusion
-
While it is likely that improvements in CoV vaccines and diagnostic tools could be achieved, due the stable CoV situation in Europe this is not currently a priority for animal health companies.
CoVs infection in pigs can cause devastating disease in naïve populations. CoVs strains that can cause massive disease outbreaks are present in many pig populations in certain geographic areas. Trade and traffic can easily promote virus transmission and introduction in distant regions at any time. Having appropriate CoV diagnostic tools in place for routine surveillance and possibly also vaccines/or the ability to upscale existing vaccines in currently CoV free areas pigs may be important.
Sources of information
-
Expert group composition
Paul Brown, French Agency for Food, Environmental and Occupational Heath Safety (ANSES), France – [Leader]
Tanja Opriessnig, The Roslin Institute, University of Edinburgh, UK and Iowa State University, USA
Sjaak De Wit, Royal GD and Veterinary Faculty of Utrecht University, Netherlands.
-
Date of submission by expert group
09-November-2020.
-
References
Alsop JE. A presumptive case of vomiting and wasting disease in a swine nucleus herd. J Swine Health Prod. 2006;14(2):97–100.
Akimkin V, Beer M, Blome S, et al. New Chimeric Porcine Coronavirus in Swine Feces, Germany, 2012. Emerg Infect Dis. 2016;22(7):1314-1315. doi:10.3201/eid2207.160179
Annamalai, T., Lin, C., Gao, X. et al. Cross protective immune responses in nursing piglets infected with a US spike-insertion deletion porcine epidemic diarrhea virus strain and challenged with an original US PEDV strain. Vet Res 48, 61 (2017).https://doi.org/10.1186/s13567-017-0469-7
Belsham GJ, Rasmussen TB, Normann P, Vaclavek P, Strandbygaard B, Bøtner A. Characterization of a Novel Chimeric Swine Enteric Coronavirus from Diseased Pigs in Central Eastern Europe in 2016. Transbound Emerg Dis. 2016;63(6):595-601. doi:10.1111/tbed.12579
Boniotti MB, Papetti A, Lavazza A, et al. Porcine Epidemic Diarrhea Virus and Discovery of a Recombinant Swine Enteric Coronavirus, Italy. Emerg Infect Dis. 2016;22(1):83-87. doi:10.3201/eid2201.150544
Bourgueil E, Hutet E, Cariolet R, Vannier P. Experimental infection of pigs with the porcine respiratory coronavirus (PRCV): measure of viral excretion. Vet Microbiol. 1992;31(1):11-18. doi:10.1016/0378-1135(92)90136-h
Brim TA, VanCott JL, Lunney JK, Saif LJ. Cellular immune responses of pigs after primary inoculation with porcine respiratory coronavirus or transmissible gastroenteritis virus and challenge with transmissible gastroenteritis virus. Vet Immunol Immunopathol. 1995;48(1-2):35-54. doi:10.1016/0165-2427(94)05416-p
Brown TT Jr. Laboratory evaluation of selected disinfectants as virucidal agents against porcine parvovirus, pseudorabies virus, and transmissible gastroenteritis virus. Am J Vet Res. 1981;42(6):1033-1036.
Brown I, Cartwright S. New porcine coronavirus?. Vet Rec. 1986;119(11):282-283. doi:10.1136/vr.119.11.282Decaro N, Lorusso A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet Microbiol. 2020 May;244:108693. doi: 10.1016/j.vetmic.2020.108693
Butler DG, Gall DG, Kelly MH, Hamilton JR. Transmissible gastroenteritis. Mechanisms responsible for diarrhea in an acute viral enteritis in piglets. J Clin Invest. 1974;53(5):1335-1342. doi:10.1172/JCI107681
Callebaut P, Enjuanes L, Pensaert M. An adenovirus recombinant expressing the spike glycoprotein of porcine respiratory coronavirus is immunogenic in swine. J Gen Virol. 1996;77 ( Pt 2 ):309-313. doi:10.1099/0022-1317-77-2-309
Cartwright SF, Harris HM, Blandford TB, Fincham I, Gitter M. A cytopathic virus causing a transmissible gastroenteritis in swine. I. Isolation and properties. J Comp Pathol. 1965;75(4):387-396. doi:10.1016/0021-9975(65)90019-8
Casanova L, Rutala WA, Weber DJ, Sobsey MD. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893-1898. doi:10.1016/j.watres.2009.02.002
Chang G, Chang T, Lin S, et al. 1993. Isolation and identification of hemagglutinating encephalomyelitis virus from pigs in Taiwan. J Chin Soc Vet Sci 19:147–158.
Chen K, He W, Lu H, et al. Development of an immunochromatographic strip for serological diagnosis of Porcine hemagglutinating encephalomyelitis virus. J Vet Diagn Invest. 2011;23(2):288-296. doi:10.1177/104063871102300214
Chen K, Zhao K, Song D, et al. Development and evaluation of an immunochromatographic strip for rapid detection of porcine hemagglutinating encephalomyelitis virus. Virol J. 2012;9:172. Published 2012 Aug 24. doi:10.1186/1743-422X-9-172
Cox E, Pensaert MB, Callebaut P. Intestinal protection against challenge with transmissible gastroenteritis virus of pigs immune after infection with the porcine respiratory coronavirus. Vaccine. 1993;11(2):267-272. doi:10.1016/0264-410x(93)90028-v
Derbyshire JB, Jessett DM, Newman G. An experimental epidemiological study of porcine transmissible gastroenteritis. J Comp Pathol. 1969;79(4):445-452. doi:10.1016/0021-9975(69)90064-4
Ding Z, Fang L, Jing H, et al. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol. 2014;88(16):8936-8945. doi:10.1128/JVI.00700-14
Dong, N., Fang, L., Zeng, S., Sun, Q., Chen, H. and Xiao, S., 2015. Porcine Deltacoronavirus in Mainland China. Emerg Infect Dis 21, 2254-5.
Frederick GT, Bohl EH. Local and systemic cell-mediated immunity against transmissible gastroenteritis, an intestinal viral infection of swine. J Immunol. 1976;116(4):1000-1004.
Gerdts V, Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol. 2017;206:45-51. doi:10.1016/j.vetmic.2016.11.029.
Gong L, Li J, Zhou Q, et al. A New Bat-HKU2-like Coronavirus in Swine, China, 2017. Emerg Infect Dis. 2017;23(9):1607-1609. doi:10.3201/eid2309.170915.
Gough PM, Jorgenson RD. Identification of porcine transmissible gastroenteritis virus in house flies (Musca domestica Linneaus). Am J Vet Res. 1983;44(11):2078-2082.
Greig AS, Bouillant AM. Studies on the hemagglutination phenomenon of hemagglutinating encephalomyelitis virus (HEV) of pigs. Can J Comp Med. 1972;36(4):366-370.
Haas B, Ahl R, Böhm R, Strauch D. Inactivation of viruses in liquid manure. Rev Sci Tech. 1995;14(2):435-445. doi:10.20506/rst.14.2.844
Haelterman EO. 1962. Epidemiological studies of transmissible gastroenteritis of swine. In United States Livestock Sanitary Association Meeting, pp. 305–315.
Hain KS, Joshi LR, Okda F, et al. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J Gen Virol. 2016;97(10):2719-2731. doi:10.1099/jgv.0.000586
Have P. Infection with a new porcine respiratory coronavirus in Denmark: serologic differentiation from transmissible gastroenteritis virus using monoclonal antibodies. Adv Exp Med Biol. 1990;276:435-439. doi:10.1007/978-1-4684-5823-7_60
Hooper BE, Haelterman EO. Growth of transmissible gastroenteritis virus in young pigs. Am J Vet Res. 1966;27(116):286-291.
Hu H, Jung K, Vlasova AN, Saif LJ. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch Virol. 2016;161(12):3421-3434. doi:10.1007/s00705-016-3056-8
Jabrane A, Girard C, Elazhary Y. Pathogenicity of porcine respiratory coronavirus isolated in Québec. Can Vet J. 1994;35(2):86-92.
Kemeny LJ, Wiltsey VL, Riley JL. Upper respiratory infection of lactating sows with transmissible gastroenteritis virus Kemeny LJ, Woods RD. Quantitative transmissible gastroenteritis virus shedding patterns in lactating sows. Am J Vet Res. 1977;38(3):307-310.
following contact exposure to infected piglets. Cornell Vet. 1975;65(3):352-362.
Killoran KE, Leedom Larson KR. Porcine respiratory coronavirus. Swine Health Information Center and Center for Food Security and Public Health, 2016. http://www.cfsph.iastate.edu/pdf/shic-factsheetporcine-respiratory-coronavirus.
Kodama Y, Ogata M, Simizu Y. Characterization of immunoglobulin A antibody in serum of swine inoculated with transmissible gastroenteritis virus. Am J Vet Res. 1980;41(5):740-745.
Lamphear BJ, Jilka JM, Kesl L, Welter M, Howard JA, Streatfield SJ. A corn-based delivery system for animal vaccines: an oral transmissible gastroenteritis virus vaccine boosts lactogenic immunity in swine. Vaccine. 2004;22(19):2420-2424. doi:10.1016/j.vaccine.2003.11.066
Laude H, Gelfi J, Aynaud JM. In vitro properties of low- and high-passaged strains of transmissible gastroenteritis coronavirus of swine. Am J Vet Res. 1981;42(3):447-449.
Laude H, Van Reeth K, Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res. 1993;24(2):125-150.
Lee C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus [published correction appears in Virol J. 2016;13:19]. Virol J. 2015;12:193. Published 2015 Dec 22. doi:10.1186/s12985-015-0421-2.
Lee DH, Jeon YS, Park CK, Kim S, Lee DS, Lee C. Immunoprophylactic effect of chicken egg yolk antibody (IgY) against a recombinant S1 domain of the porcine epidemic diarrhea virus spike protein in piglets. Arch Virol. 2015 Sep;160(9):2197-207. doi: 10.1007/s00705-015-2494-z. Epub 2015 Jun 23.
Lee DU, Kwon T, Je SH, et al. Wild boars harboring porcine epidemic diarrhea virus (PEDV) may play an important role as a PEDV reservoir. Vet Microbiol. 2016;192:90-94. doi:10.1016/j.vetmic.2016.07.003
Li Z, He W, Lan Y, et al. The evidence of porcine hemagglutinating encephalomyelitis virus induced nonsuppurative encephalitis as the cause of death in piglets. PeerJ. 2016;4:e2443. Published 2016 Sep 15. doi:10.7717/peerj.2443
Li X, Wang L, Zhen Y, Li S, Xu Y. Chicken egg yolk antibodies (IgY) as non-antibiotic production enhancers for use in swine production: a review. J Anim Sci Biotechnol. 2015 Aug 25;6(1):40. doi: 10.1186/s40104-015-0038-8.
Liu DQ, Ge JW, Qiao XY, Jiang YP, Liu SM, Li YJ. High-level mucosal and systemic immune responses induced by oral administration with Lactobacillus-expressed porcine epidemic diarrhea virus (PEDV) S1 region combined with Lactobacillus-expressed N protein. Appl Microbiol Biotechnol. 2012;93(6):2437-2446. doi:10.1007/s00253-011-3734-0
Liu X, Zhao D, Zhou P, Zhang Y, Wang Y. Evaluation of the Efficacy of a Recombinant Adenovirus Expressing the Spike Protein of Porcine Epidemic Diarrhea Virus in Pigs. Biomed Res Int. 2019;2019:8530273. Published 2019 Oct 8. doi:10.1155/2019/8530273
Ma Y, Zhang Y, Liang X, et al. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6(2):e00064. Published 2015 Mar 10. doi:10.1128/mBio.00064-15
McClurkin AW, Stark SL, Norman JO. Transmissible gastroenteritis (TGE) of swine: the possible role of dogs in the epizootiology of TGE. Can J Comp Med. 1970;34(4):347-349.
Moon HW. Mechanisms in the pathogenesis of diarrhea: a review. J Am Vet Med Assoc. 1978;172(4):443-448.
Mora-Díaz JC, Piñeyro PE, Houston E, Zimmerman J, Giménez-Lirola LG. Porcine Hemagglutinating Encephalomyelitis Virus: A Review. Front Vet Sci. 2019;6:53. Published 2019 Feb 27. doi:10.3389/fvets.2019.00053
Mora-Díaz JC, Magtoto R, Houston E, et al. Detecting and Monitoring Porcine Hemagglutinating Encephalomyelitis Virus, an Underresearched Betacoronavirus. mSphere. 2020;5(3):e00199-20. Published 2020 May 6. doi:10.1128/mSphere.00199-20
Niederwerder MC, Nietfeld JC, Bai J, et al. Tissue localization, shedding, virus carriage, antibody response, and aerosol transmission of Porcine epidemic diarrhea virus following inoculation of 4-week-old feeder pigs. J Vet Diagn Invest. 2016;28(6):671-678. doi:10.1177/1040638716663251
OIE Technical Factsheet: Infection with porcine epidemic diarrhoea virus. https://www.oie.int/doc/ged/D13924.PDF
Onno M, Jestin A, Cariolet R, Vannier P. Rapid diagnosis of TGEV-like coronavirus in fattened pigs by indirect immunofluorescence labelling in nasal cells. Zentralbl Veterinarmed B. 1989;36(8):629-634. doi:10.1111/j.1439-0450.1989.tb00654.x
Pan Y, Tian X, Qin P, et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol. 2017;211:15-21. doi:10.1016/j.vetmic.2017.09.020
Paul PS, Mengeling WL. Persistence of passively acquired antibodies to hemagglutinating encephalomyelitis virus in swine. Am J Vet Res. 1984;45(5):932-934.
Pensert M,. Overview of porcine hemaggluinating encephalomyelitis. 213. MSD Manualhttps://www.msdvetmanual.com/generalized-conditions/porcine-hemagglutinating-encephalomyelitis/overview-of-porcine-hemagglutinating-encephalomyelitis
Pensaert MB, Callebaut PE. Characteristics of a coronavirus causing vomition and wasting in pigs. Arch Gesamte Virusforsch. 1974;44(1):35-50. doi:10.1007/BF01242179
Pensaert M, Callebaut P, Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q. 1986;8(3):257-261. doi:10.1080/01652176.1986.9694050
Pensaert M, Cox E, van Deun K, Callebaut P. A sero-epizootiological study of porcine respiratory coronavirus in Belgian swine. Vet Q. 1993;15(1):16-20. doi:10.1080/01652176.1993.9694361
Penzes Z, Gonzalez JM, Calvo E, et al. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes. 2001;23(1):105-118. doi:10.1023/a:1011147832586
Pilchard EI. Experimental transmission of transmissible gastroenteritis virus by starlings. Am J Vet Res. 1965;26(114):1177-1179.
Reynolds DJ, Garwes DJ. Virus isolation and serum antibody responses after infection of cats with transmissible gastroenteritis virus. Brief report. Arch Virol. 1979;60(2):161-166. doi:10.1007/BF01348032
Saif LJ, Bohl EH. Passive immunity to transmissible gastroenteritis virus: intramammary viral inoculation of sows. Ann N Y Acad Sci. 1983;409:708-723. doi:10.1111/j.1749-6632.1983.tb26910.x
Saif LJ, van Cott JL, Brim TA. Immunity to transmissible gastroenteritis virus and porcine respiratory coronavirus infections in swine. Vet Immunol Immunopathol. 1994;43(1-3):89-97. doi:10.1016/0165-2427(94)90124-4
Saif LJ, Wang Q, Vlasova AN, Jung K, Xiao S. Chapter 31: Coronaviruses. p488-523. In: Diseases of Swine, Eleventh Edition. Edited by Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KL, Stevenson GW, Zhang J. © 2019 John Wiley & Sons, Inc. Published 2019 by John Wiley & Sons, Inc.
Schulz LL, Tonsor GT. Assessment of the economic impacts of porcine epidemic diarrhea virus in the United States. J Anim Sci. 2015;93(11):5111-5118. doi:10.2527/jas.2015-9136.
Sedlak K, Bartova E, Machova J. Antibodies to selected viral disease agents in wild boars from the Czech Republic. J Wildl Dis. 2008;44(3):777-780. doi:10.7589/0090-3558-44.3.777
Shimizu M, Shimizu Y. Demonstration of cytotoxic lymphocytes to virus-infected target cells in pigs inoculated with transmissible gastroenteritis virus. Am J Vet Res. 1979;40(2):208-213.
Song, D., Zhou, X., Peng, Q., Chen, Y., Zhang, F., Huang, T., Zhang, T., Li, A., Huang, D., Wu, Q., He, H. and Tang, Y., 2015. Newly Emerged Porcine Deltacoronavirus Associated With Diarrhoea in Swine in China: Identification, Prevalence and Full-Length Genome Sequence Analysis. Transbound Emerg Dis 62, 575-80.
Stevenson GW, Hoang H, Schwartz KJ, et al. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25(5):649-654. doi:10.1177/1040638713501675Zhou P, Fan H, Lan T, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255-258. doi:10.1038/s41586-018-0010-9.
TGE ISU Swine Manual. https://vetmed.iastate.edu/vdpam/FSVD/swine/index-diseases/tge
Underdahl NR, Mebus CA, Torres-Medina A. Recovery of transmissible gastroenteritis virus from chronically infected experimental pigs. Am J Vet Res. 1975;36(10):1473-1476.
Usami Y, Fukai K, Ichikawa Y, et al. Virological and serological studies of porcine respiratory coronavirus infection on a Japanese farm. J Vet Med Sci. 2008;70(9):929-936. doi:10.1292/jvms.70.929
VanCott JL, Brim TA, Simkins RA, Saif LJ. Isotype-specific antibody-secreting cells to transmissible gastroenteritis virus and porcine respiratory coronavirus in gut- and bronchus-associated lymphoid tissues of suckling pigs. J Immunol. 1993;150(9):3990-4000.
VanCott JL, Brim TA, Lunney JK, Saif LJ. Contribution of antibody-secreting cells induced in mucosal lymphoid tissues of pigs inoculated with respiratory or enteric strains of coronavirus to immunity against enteric coronavirus challenge. J Immunol. 1994;152(8):3980-3990.
Vaughn EM, Halbur PG, Paul PS. Three new isolates of porcine respiratory coronavirus with various pathogenicities and spike (S) gene deletions. J Clin Microbiol. 1994;32(7):1809-1812.
Vaughn EM, Halbur PG, Paul PS. Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J Virol. 1995;69(5):3176-3184.
Vlasova A.N., Wang Q., Jung K., Langel S.N., Malik Y.S., Saif L.J. (2020) Porcine Coronaviruses. In: Malik Y., Singh R., Yadav M. (eds) Emerging and Transboundary Animal Viruses. Livestock Diseases and Management. Springer, Singapore
Wang D, Fang L, Shi Y, et al. Porcine Epidemic Diarrhea Virus 3C-Like Protease Regulates Its Interferon Antagonism by Cleaving NEMO. J Virol. 2015;90(4):2090-2101. Published 2015 Dec 9. doi:10.1128/JVI.02514-15
Wang, Q., Vlasova, A.N., Kenney, S.P. and Saif, L.J., 2019. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol 34, 39-49.
Wesley RD, Woods RD. Induction of protective immunity against transmissible gastroenteritis virus after exposure of neonatal pigs to porcine respiratory coronavirus. Am J Vet Res. 1996;57(2):157-162.
Wesley RD, Woods RD, McKean JD, Senn MK, Elazhary Y. Prevalence of coronavirus antibodies in Iowa swine. Can J Vet Res. 1997;61(4):305-308.
Wesley RD, Woods RD, Hill HT, Biwer JD. Evidence for a porcine respiratory coronavirus, antigenically similar to transmissible gastroenteritis virus, in the United States. J Vet Diagn Invest. 1990;2(4):312-317. doi:10.1177/104063879000200411
Woods RD, Wesley RD. Transmissible gastroenteritis coronavirus carrier sow. Adv Exp Med Biol. 1998;440:641-647. doi:10.1007/978-1-4615-5331-1_83
Yang YL, Yu JQ, Huang YW. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. 2020;285:198024. doi:10.1016/j.virusres.2020.198024
Zhang J. Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71-84. doi:10.1016/j.virusres.2016.05.028
Zhang Y, Cheng Y, Xing G, et al. Detection and spike gene characterization in porcine deltacoronavirus in China during 2016-2018. Infect Genet Evol. 2019;73:151-158. doi:10.1016/j.meegid.2019.04.023
Zhang X, Hasoksuz M, Spiro D, et al. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358(2):424-435. doi:10.1016/j.virol.2006.08.051
Zhang Y, Zhang X, Liao X, et al. Construction of a bivalent DNA vaccine co-expressing S genes of transmissible gastroenteritis virus and porcine epidemic diarrhea virus delivered by attenuated Salmonella typhimurium. Virus Genes. 2016;52(3):354-364. doi:10.1007/s11262-016-1316-z.
Zhang X, Hasoksuz M, Spiro D, et al. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358(2):424-435. doi:10.1016/j.virol.2006.08.051
Zhang X, Zhu Y, Zhu X, et al. Identification of a natural recombinant transmissible gastroenteritis virus between Purdue and Miller clusters in China. Emerg Microbes Infect. 2017;6(8):e74. Published 2017 Aug 23. doi:10.1038/emi.2017.62
Zhang Q, Yoo D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviralinnate signaling. Virus Res. 2016;226:128-141. doi:10.1016/j.virusres.2016.05.015
